Methionine
 Skeletal formula of the canonical form of methionine
| |||
| |||
| Names | |||
|---|---|---|---|
| IUPAC name
Methionine
| |||
| Other names
2-amino-4-(methylthio)butanoic acid
| |||
| Identifiers | |||
3D model (JSmol)
|
|||
| Abbreviations | Met, M | ||
| ChEBI |
| ||
| ChEMBL |
| ||
| ChemSpider | |||
| DrugBank |
| ||
| ECHA InfoCard | 100.000.393 | ||
| EC Number |
| ||
| KEGG |
| ||
PubChem CID
|
|||
| UNII |
| ||
CompTox Dashboard (EPA)
|
|||
| |||
| |||
| Properties[2] | |||
| C5H11NO2S | |||
| Molar mass | 149.21 g·mol−1 | ||
| Appearance | White crystalline powder | ||
| Density | 1.340 g/cm3 | ||
| Melting point | 281 °C (538 °F; 554 K) decomposes | ||
| Soluble | |||
| Acidity (pKa) | 2.28 (carboxyl), 9.21 (amino)[1] | ||
| Pharmacology | |||
| V03AB26 (WHO) QA05BA90 (WHO), QG04BA90 (WHO) | |||
| Supplementary data page | |||
| Methionine (data page) | |||
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |||
Methionine (symbol Met or M)[3] (/mɪˈθaɪəniːn/)[4] is an essential amino acid in humans.
As the precursor of other non-essential amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical role in the metabolism and health of many species, including humans. Methionine is also involved in angiogenesis and various processes related to DNA transcription, epigenetic expression, and gene regulation.
Methionine was first isolated in 1921 by John Howard Mueller.[5] It is encoded by the codon AUG. It was named by Satoru Odake in 1925, as an abbreviation of its structural description 2-amino-4-(methylthio)butanoic acid.[6]
Biochemical details
[edit]Methionine (abbreviated as Met or M; encoded by the codon AUG) is an α-amino acid that is used in the biosynthesis of proteins. It contains a carboxyl group (which is in the deprotonated −COO− form under biological pH conditions), an amino group (which is in the protonated −NH+
3 form under biological pH conditions) located in α-position with respect to the carboxyl group, and an S-methyl thioether side chain, classifying it as a nonpolar, aliphatic amino acid.[citation needed]
In nuclear genes of eukaryotes and in Archaea, methionine is coded for by the start codon, meaning it indicates the start of the coding region and is the first amino acid produced in a nascent polypeptide during mRNA translation.[7]
A proteinogenic amino acid
[edit]Cysteine and methionine are the two sulfur-containing proteinogenic amino acids. Excluding the few exceptions where methionine may act as a redox sensor (e.g.,methionine sulfoxide[8]), methionine residues do not have a catalytic role.[9] This is in contrast to cysteine residues, where the thiol group has a catalytic role in many proteins.[9] The thioether within methionine does however have a minor structural role due to the stability effect of S/π interactions between the side chain sulfur atom and aromatic amino acids in one-third of all known protein structures.[9] This lack of a strong role is reflected in experiments where little effect is seen in proteins where methionine is replaced by norleucine, a straight hydrocarbon sidechain amino acid which lacks the thioether.[10] It has been conjectured that norleucine was present in early versions of the genetic code, but methionine intruded into the final version of the genetic code due to the fact it is used in the cofactor S-adenosylmethionine (SAM-e).[11] This situation is not unique and may have occurred with ornithine and arginine.[12]
Encoding
[edit]Methionine is one of only two amino acids encoded by a single codon (AUG) in the standard genetic code (tryptophan, encoded by UGG, is the other). In reflection to the evolutionary origin of its codon, the other AUN codons encode isoleucine, which is also a hydrophobic amino acid. In the mitochondrial genome of several organisms, including metazoa and yeast, the codon AUA also encodes for methionine. In the standard genetic code AUA codes for isoleucine and the respective tRNA (ileX in Escherichia coli) uses the unusual base lysidine (bacteria) or agmatidine (archaea) to discriminate against AUG.[13][14]
The methionine codon AUG is also the most common start codon. A "Start" codon is message for a ribosome that signals the initiation of protein translation from mRNA when the AUG codon is in a Kozak consensus sequence. As a consequence, methionine is often incorporated into the N-terminal position of proteins in eukaryotes and archaea during translation, although it can be removed by post-translational modification. In bacteria, the derivative N-formylmethionine is used as the initial amino acid.[citation needed]
Derivatives
[edit]S-Adenosylmethionine
[edit]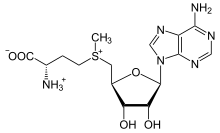
The methionine-derivative S-adenosylmethionine (SAM-e) is a cofactor that serves mainly as a methyl donor. SAM-e is composed of an adenosyl molecule (via 5′ carbon) attached to the sulfur of methionine, therefore making it a sulfonium cation (i.e., three substituents and positive charge). The sulfur acts as a soft Lewis acid (i.e., donor/electrophile) which allows the S-methyl group to be transferred to an oxygen, nitrogen, or aromatic system, often with the aid of other cofactors such as cobalamin (vitamin B12 in humans). Some enzymes use SAM-e to initiate a radical reaction; these are called radical SAM-e enzymes. As a result of the transfer of the methyl group, S-adenosylhomocysteine is obtained. In bacteria, this is either regenerated by methylation or is salvaged by removing the adenine and the homocysteine, leaving the compound dihydroxypentandione to spontaneously convert into autoinducer-2, which is excreted as a waste product or quorum signal.[citation needed]
Biosynthesis
[edit]
As an essential amino acid, methionine is not synthesized de novo in humans and other animals, which must ingest methionine or methionine-containing proteins. In plants and microorganisms, methionine biosynthesis belongs to the aspartate family, along with threonine and lysine (via diaminopimelate, but not via α-aminoadipate). The main backbone is derived from aspartic acid, while the sulfur may come from cysteine, methanethiol, or hydrogen sulfide.[9]
- First, aspartic acid is converted via β-aspartyl semialdehyde into homoserine by two reduction steps of the terminal carboxyl group (homoserine has therefore a γ-hydroxyl, hence the homo- series). The intermediate aspartate semialdehyde is the branching point with the lysine biosynthetic pathway, where it is instead condensed with pyruvate. Homoserine is the branching point with the threonine pathway, where instead it is isomerised after activating the terminal hydroxyl with phosphate (also used for methionine biosynthesis in plants).[9]
- Homoserine is then activated with a phosphate, succinyl or an acetyl group on the hydroxyl.
- In plants and possibly in some bacteria,[9] phosphate is used. This step is shared with threonine biosynthesis.[9]
- In most organisms, an acetyl group is used to activate the homoserine. This can be catalysed in bacteria by an enzyme encoded by metX or metA (not homologues).[9]
- In enterobacteria and a limited number of other organisms, succinate is used. The enzyme that catalyses the reaction is MetA and the specificity for acetyl-CoA and succinyl-CoA is dictated by a single residue.[9] The physiological basis for the preference of acetyl-CoA or succinyl-CoA is unknown, but such alternative routes are present in some other pathways (e.g. lysine biosynthesis and arginine biosynthesis).
- The hydroxyl activating group is then replaced with cysteine, methanethiol, or hydrogen sulfide. A replacement reaction is technically a γ-elimination followed by a variant of a Michael addition. All the enzymes involved are homologues and members of the Cys/Met metabolism PLP-dependent enzyme family, which is a subset of the PLP-dependent fold type I clade. They utilise the cofactor PLP (pyridoxal phosphate), which functions by stabilising carbanion intermediates.[9]
- If it reacts with cysteine, it produces cystathionine, which is cleaved to yield homocysteine. The enzymes involved are cystathionine-γ-synthase (encoded by metB in bacteria) and cystathionine-β-lyase (metC). Cystathionine is bound differently in the two enzymes allowing β or γ reactions to occur.[9]
- If it reacts with free hydrogen sulfide, it produces homocysteine. This is catalysed by O-acetylhomoserine aminocarboxypropyltransferase (formerly known as O-acetylhomoserine (thiol)-lyase. It is encoded by either metY or metZ in bacteria.[9]
- If it reacts with methanethiol, it produces methionine directly. Methanethiol is a byproduct of catabolic pathway of certain compounds, therefore this route is more uncommon.[9]
- If homocysteine is produced, the thiol group is methylated, yielding methionine. Two methionine synthases are known; one is cobalamin (vitamin B12) dependent and one is independent.[9]
The pathway using cysteine is called the "transsulfuration pathway", while the pathway using hydrogen sulfide (or methanethiol) is called "direct-sulfurylation pathway".
Cysteine is similarly produced, namely it can be made from an activated serine and either from homocysteine ("reverse transsulfurylation route") or from hydrogen sulfide ("direct sulfurylation route"); the activated serine is generally O-acetylserine (via CysK or CysM in E. coli), but in Aeropyrum pernix and some other archaea O-phosphoserine is used.[15] CysK and CysM are homologues, but belong to the PLP fold type III clade.[citation needed]
Transsulfurylation pathway
[edit]Enzymes involved in the E. coli transsulfurylation route of methionine biosynthesis:[citation needed]
- Aspartokinase
- Aspartate-semialdehyde dehydrogenase
- Homoserine dehydrogenase
- Homoserine O-transsuccinylase
- Cystathionine-γ-synthase
- Cystathionine-β-lyase
- Methionine synthase (in mammals, this step is performed by homocysteine methyltransferase or betaine—homocysteine S-methyltransferase.)
Other biochemical pathways
[edit]
Although mammals cannot synthesize methionine, they can still use it in a variety of biochemical pathways:
Catabolism
[edit]Methionine is converted to S-adenosylmethionine (SAM-e) by (1) methionine adenosyltransferase.[citation needed]
SAM-e serves as a methyl donor in many (2) methyltransferase reactions, and is converted to S-adenosylhomocysteine (SAH).[citation needed]
(3) Adenosylhomocysteinase cysteine.
Regeneration
[edit]Methionine can be regenerated from homocysteine via (4) methionine synthase in a reaction that requires vitamin B12 as a cofactor.[citation needed]
Homocysteine can also be remethylated using glycine betaine (N,N,N-trimethylglycine, TMG) to methionine via the enzyme betaine-homocysteine methyltransferase (E.C.2.1.1.5, BHMT). BHMT makes up to 1.5% of all the soluble protein of the liver, and recent evidence suggests that it may have a greater influence on methionine and homocysteine homeostasis than methionine synthase.[citation needed]
Reverse-transulfurylation pathway: conversion to cysteine
[edit]Homocysteine can be converted to cysteine.
- (5) Cystathionine-β-synthase (an enzyme which requires pyridoxal phosphate, the active form of vitamin B6) combines homocysteine and serine to produce cystathionine. Instead of degrading cystathionine via cystathionine-β-lyase, as in the biosynthetic pathway, cystathionine is broken down to cysteine and α-ketobutyrate via (6) cystathionine-γ-lyase.[citation needed]
- (7) The enzyme α-ketoacid dehydrogenase converts α-ketobutyrate to propionyl-CoA, which is metabolized to succinyl-CoA in a three-step process (see propionyl-CoA for pathway).[citation needed]
Ethylene synthesis
[edit]This amino acid is also used by plants for synthesis of ethylene. The process is known as the Yang cycle or the methionine cycle.
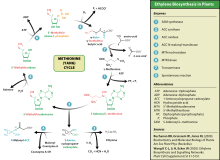
Metabolic diseases
[edit]The degradation of methionine is impaired in the following metabolic diseases:[citation needed]
- Combined malonic and methylmalonic aciduria (CMAMMA)
- Homocystinuria
- Methylmalonic acidemia
- Propionic acidemia
Chemical synthesis
[edit]The industrial synthesis combines acrolein, methanethiol, and cyanide, which affords the hydantoin.[16] Racemic methionine can also be synthesized from diethyl sodium phthalimidomalonate by alkylation with chloroethylmethylsulfide (ClCH2CH2SCH3) followed by hydrolysis and decarboxylation. Also see Methanol. [17]
Human nutrition
[edit]There is inconclusive clinical evidence on methionin supplementation.[18] Dietary restriction of methionine can lead to bone-related disorders.[18]
Methionine supplementation may benefit those suffering from copper poisoning.[19]
Overconsumption of methionine, the methyl group donor in DNA methylation, is related to cancer growth in a number of studies.[20][21]
Requirements
[edit]The Food and Nutrition Board of the U.S. Institute of Medicine set Recommended Dietary Allowances (RDAs) for essential amino acids in 2002. For methionine combined with cysteine, for adults 19 years and older, 19 mg/kg body weight/day.[22]
This translates to about 1.33 grams per day for a 70 kilogram individual.[citation needed]
Dietary sources
[edit]| Food | g/100 g |
|---|---|
| Egg, white, dried, powder, glucose reduced | 3.204 |
| Sesame seeds flour (low fat) | 1.656 |
| Brazil nuts | 1.124 |
| Cheese, Parmesan, shredded | 1.114 |
| hemp seed, hulled | 0.933 |
| Soy protein concentrate | 0.814 |
| Chicken, broilers or fryers, roasted | 0.801 |
| Fish, tuna, light, canned in water, drained solids | 0.755 |
| Beef, cured, dried | 0.749 |
| Bacon | 0.593 |
| chia seeds | 0.588 |
| Beef, ground, 95% lean meat / 5% fat, raw | 0.565 |
| Pork, ground, 96% lean / 4% fat, raw | 0.564 |
| Soybeans | 0.547 |
| Wheat germ | 0.456 |
| Egg, whole, cooked, hard-boiled | 0.392 |
| Oat | 0.312 |
| Peanuts | 0.309 |
| Chickpea | 0.253 |
| Corn, yellow | 0.197 |
| Almonds | 0.151 |
| Beans, pinto, cooked | 0.117 |
| Lentils, cooked | 0.077 |
| Rice, brown, medium-grain, cooked | 0.052 |
High levels of methionine can be found in eggs, meat, and fish; sesame seeds, Brazil nuts, and some other plant seeds; and cereal grains. Most fruits and vegetables contain very little. Most legumes, though protein dense, are low in methionine. Proteins without adequate methionine are not considered to be complete proteins.[23] For that reason, racemic methionine is sometimes added as an ingredient to pet foods.[24]
Health
[edit]Loss of methionine has been linked to senile greying of hair. Its lack leads to a buildup of hydrogen peroxide in hair follicles, a reduction in tyrosinase effectiveness, and a gradual loss of hair color.[25] Methionine raises the intracellular concentration of glutathione, thereby promoting antioxidant-mediated cell defense and redox regulation. It also protects cells against dopamine induced nigral cell loss by binding oxidative metabolites.[26]
Methionine is an intermediate in the biosynthesis of cysteine, carnitine, taurine, lecithin, phosphatidylcholine, and other phospholipids. Improper conversion of methionine can lead to atherosclerosis[27] due to accumulation of homocysteine.
Other uses
[edit]DL-Methionine is sometimes given as a supplement to dogs; It helps reduce the chances of kidney stones in dogs. Methionine is also known to increase the urinary excretion of quinidine by acidifying the urine. Aminoglycoside antibiotics used to treat urinary tract infections work best in alkaline conditions, and urinary acidification from using methionine can reduce its effectiveness. If a dog is on a diet that acidifies the urine, methionine should not be used.[28]
Methionine is allowed as a supplement to organic poultry feed under the US certified organic program.[29]
Methionine can be used as a nontoxic pesticide option against giant swallowtail caterpillars, which are a serious pest to orange crops.[30]
See also
[edit]- Allantoin
- Formylmethionine
- Methionine oxidation
- Paracetamol poisoning
- Photoreactive methionine
- S-Methylcysteine
References
[edit]- ^ Dawson RM, Elliott DC, Elliott WH, Jones KM (1959). Data for Biochemical Research. Oxford: Clarendon Press.
- ^ Weast, Robert C., ed. (1981). CRC Handbook of Chemistry and Physics (62nd ed.). Boca Raton, FL: CRC Press. p. C-374. ISBN 0-8493-0462-8..
- ^ "Nomenclature and Symbolism for Amino Acids and Peptides". IUPAC-IUB Joint Commission on Biochemical Nomenclature. 1983. Archived from the original on 9 October 2008. Retrieved 5 March 2018.
- ^ "Methionine". Oxford University Press. Archived from the original on January 27, 2018.
- ^ Pappenheimer AM (1987). "A Biographical Memoir of John Howard Mueller" (PDF). Washington D.C.: National Academy of Sciences.
- ^ Odake, Satoru (1925). "On the Occurrence of a Sulphur-containing Amino acid in Yeast". Bulletin of the Agricultural Chemical Society of Japan. 1 (8): 87–89. doi:10.1271/bbb1924.1.87. ISSN 1881-1272.
- ^ Guedes RL, Prosdocimi F, Fernandes GR, Moura LK, Ribeiro HA, Ortega JM (December 2011). "Amino acids biosynthesis and nitrogen assimilation pathways: a great genomic deletion during eukaryotes evolution". BMC Genomics. 12 (Suppl 4): S2. doi:10.1186/1471-2164-12-S4-S2. PMC 3287585. PMID 22369087.
- ^ Bigelow DJ, Squier TC (January 2005). "Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics (Submitted manuscript). 1703 (2): 121–134. doi:10.1016/j.bbapap.2004.09.012. PMID 15680220.
- ^ a b c d e f g h i j k l m n Ferla MP, Patrick WM (August 2014). "Bacterial methionine biosynthesis". Microbiology. 160 (Pt 8): 1571–1584. doi:10.1099/mic.0.077826-0. PMID 24939187.
- ^ Cirino PC, Tang Y, Takahashi K, Tirrell DA, Arnold FH (September 2003). "Global incorporation of norleucine in place of methionine in cytochrome P450 BM-3 heme domain increases peroxygenase activity". Biotechnology and Bioengineering. 83 (6): 729–734. doi:10.1002/bit.10718. PMID 12889037. S2CID 11380413.
- ^ Alvarez-Carreño C, Becerra A, Lazcano A (October 2013). "Norvaline and norleucine may have been more abundant protein components during early stages of cell evolution". Origins of Life and Evolution of the Biosphere. 43 (4–5): 363–375. Bibcode:2013OLEB...43..363A. doi:10.1007/s11084-013-9344-3. PMID 24013929. S2CID 17224537.
- ^ Jukes TH (August 1973). "Arginine as an evolutionary intruder into protein synthesis". Biochemical and Biophysical Research Communications. 53 (3): 709–714. doi:10.1016/0006-291x(73)90151-4. PMID 4731949.
- ^ Ikeuchi Y, Kimura S, Numata T, Nakamura D, Yokogawa T, Ogata T, Wada T, Suzuki T, Suzuki T (April 2010). "Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea". Nature Chemical Biology. 6 (4): 277–282. doi:10.1038/nchembio.323. PMID 20139989.
- ^ Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S (November 1988). "Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification". Nature. 336 (6195): 179–181. Bibcode:1988Natur.336..179M. doi:10.1038/336179a0. PMID 3054566. S2CID 4371485.
- ^ Mino K, Ishikawa K (September 2003). "A novel O-phospho-L-serine sulfhydrylation reaction catalyzed by O-acetylserine sulfhydrylase from Aeropyrum pernix K1". FEBS Letters. 551 (1–3): 133–138. doi:10.1016/S0014-5793(03)00913-X. PMID 12965218. S2CID 28360765.
- ^ Karlheinz Drauz; Ian Grayson; Axel Kleemann; Hans-Peter Krimmer; Wolfgang Leuchtenberger; Christoph Weckbecker (2006). Ullmann's Encyclopedia of Industrial Chemistry. Weinheim: Wiley-VCH. doi:10.1002/14356007.a02_057.pub2. ISBN 978-3527306732.
- ^ Barger G, Weichselbaum TE (1934). "dl-Methionine". Organic Syntheses. 14: 58; Collected Volumes, vol. 2, p. 384.
- ^ "Methionine". WebMD.
- ^ Cavuoto P, Fenech MF (2012). "A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension". Cancer Treatment Reviews. 38 (6): 726–736. doi:10.1016/j.ctrv.2012.01.004. PMID 22342103.
- ^ Cellarier E, Durando X, Vasson MP, Farges MC, Demiden A, Maurizis JC, Madelmont JC, Chollet P (2003). "Methionine dependency and cancer treatment". Cancer Treatment Reviews. 29 (6): 489–499. doi:10.1016/S0305-7372(03)00118-X. PMID 14585259.
- ^ Institute of Medicine (2002). "Protein and Amino Acids". Dietary Reference Intakes for Energy, Carbohydrates, Fiber, Fat, Fatty Acids, Cholesterol, Protein, and Amino Acids. Washington, DC: The National Academies Press. pp. 589–768. doi:10.17226/10490. ISBN 978-0-309-08525-0.
- ^ Finkelstein JD (May 1990). "Methionine metabolism in mammals". The Journal of Nutritional Biochemistry. 1 (5): 228–237. doi:10.1016/0955-2863(90)90070-2. PMID 15539209. S2CID 32264340.
- ^ Palika L (1996). The Consumer's Guide to Dog Food: What's in Dog Food, Why It's There and How to Choose the Best Food for Your Dog. New York: Howell Book House. ISBN 978-0-87605-467-3.
- ^ Wood JM, Decker H, Hartmann H, Chavan B, Rokos H, Spencer JD, et al. (July 2009). "Senile hair graying: H2O2-mediated oxidative stress affects human hair color by blunting methionine sulfoxide repair". FASEB Journal. 23 (7): 2065–75. arXiv:0706.4406. doi:10.1096/fj.08-125435. hdl:10454/6241. PMID 19237503. S2CID 16069417.
- ^ Pinnen F, et al. (2009). "Codrugs linking L-dopa and sulfur-containing antioxidants: new pharmacological tools against Parkinson's disease". Journal of Medicinal Chemistry. 52 (2): 559–63. doi:10.1021/jm801266x. PMID 19093882.
- ^ Refsum H, Ueland PM, Nygård O, Vollset SE (1998). "Homocysteine and cardiovascular disease". Annual Review of Medicine. 49 (1): 31–62. doi:10.1146/annurev.med.49.1.31. PMID 9509248.
- ^ Grimshaw, Jane (July 25, 2011) Methionine for Dogs uses and Side Effects. critters360.com
- ^ "Rules and Regulations". Federal Register. 76 (49): 13501–13504. March 14, 2011.
- ^ Lewis DS, Cuda JP, Stevens BR (December 2011). "A novel biorational pesticide: efficacy of methionine against Heraclides (Papilio) cresphontes, a surrogate of the invasive Princeps (Papilio) demoleus (Lepidoptera: Papilionidae)". Journal of Economic Entomology. 104 (6): 1986–1990. doi:10.1603/ec11132. PMID 22299361. S2CID 45255198.
External links
[edit]- Rudra MN, Chowdhury LM (30 September 1950). "Methionine Content of Cereals and Legumes". Nature. 166 (568): 568. Bibcode:1950Natur.166..568R. doi:10.1038/166568a0. PMID 14780151. S2CID 3026278.



